

Index |
Last Chapter |
Next Chapter |


In the last fifty years electronic computers have come from being a military curiosity to a ubiquitous tool. In the same way as the lever and the wheel once extended the physical power of human beings, computers have been able to extend our mental powers, allowing us to process huge quantities of information in hitherto undreamt of ways.
For the researcher, it is becoming possible to examine the world using entirely new tools, making sense of large quantities of data where before we would have been overwhelmed by the task. It is also becoming possible for us to ask complex "what if?" questions about the world, and answer them with something more than pure supposition and imagination. Perhaps the most amazing thing about computers is not that they perform calculations at blinding speeds, or that they can communicate with each other at an incredible rate, but that they allow us to visualize, model and, in a way, interact with environments that would never otherwise be possible.
Through computer modelling we can create virtual environments that allow us to examine the entirety of creation, from galaxies and nebulas to atoms and molecules. We can even attempt to model the processes that occur within a living cell.
In the sciences, two of the most valuable things that computers can be used for are improving the analysis of data, and simulating experiments and conditions that are either impossible, impractical, or too expensive to perform physically. Often computers make procedures practical that would otherwise have required large teams of people with slide rules or complex equipment, or might have been completely impossible.
However, one of the current problems with computers is that it is difficult for non-experts to use computers as effectively as possible, especially in specialized fields where pre-written software may not be available. This has led to a number of collaborations, where a specialist in one field works with a computer-literate non-specialist, in an attempt to apply computers to a particular field.
This thesis represents such a collaboration, between plant cell physiologists and computer scientists, initially to examine the transport of material through the tunnels that join plant cells. However, this led into the wider area of transport in general, and from there into a much broader field altogether.
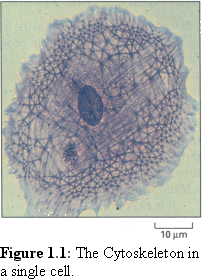
The cytoskeleton (literally, 'skeleton of the cell') is a complex,
dynamic set of structural components within a cell that give the
cell shape, and act as an internal 'railroad' for the internal
movement of cell components. Unfortunately the cytoskeleton
is difficult to observe in action. The filaments
(Fig. 1.1 (1)) of the
cytoskeleton are too small to observe in detail by light
microscopy (although much interesting work has been done on
the aggregated filaments using fluorescence microscopy). Nor
can they be observed in detail with an electron microscope
without killing the cell. Another complication is that this
system of filaments is dynamic, and is continuously being
destroyed and recreated within the living cell.

While the static structure of these cytoskeletal components is
quite well known, many elements involved in their dynamic
operation are poorly understood. Actin filaments and
microtubules (cf Fig. 1.2), which are two of the main
components of the cytoskeleton, are usually found in a cyclic
state of growth and decay, changing their lengths rapidly
depending on various environmental conditions that are not
fully understood. They also act in concert with movement
proteins such as myosin and dynein, which in turn carry or
move other proteins around the cell.
These processes are not well understood either.
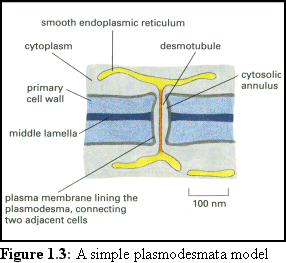
The cytoskeleton is responsible for movement
within both animal and plant cells. Movement
between cells is mediated by a number of
different structures, the largest of which are
gap junctions in animal cells, and
plasmodesmata in plant cells. Animal cells (in
general) are surrounded by a thin, flexible
membrane, bridged by the gap junction. Plant
cells however have a thick cellulose cell wall,
and the plasmodesmata that pass through them
take the form of long tunnels (see Fig. 1.3 (2)).
However, like cytoskeletal filaments however, plasmodesmata are too small to observe using light microscopy. Their internal structure is also very difficult to ascertain using current electron micrographic techniques. Plasmodesmata are so small, and their internal structures so tightly integrated into their surrounding membranes, that they are difficult to separate from the cell wall in which they are embedded, making it hard to prepare specimens for high power electron microscopy. Being embedded in the cell wall makes it difficult to get a clean cross-sectional cut, and also impairs isolation of their compositional proteins. In addition, as cellular gateways they are very sensitive to injury, and will react to close themselves if they sense any injury, as frequently happens during specimen preparation (3).
Direct protein analysis is problematic, since it is hard to prepare a large enough quantity of the plasmodesmal proteins independent of the surrounding cell wall proteins, although some attempts have been made (4), (5). One approach that has been successful is using antibodies to known proteins and examining whether they bind to active sites on exposed proteins in plasmodesmata (6). Using antibodies, it has been shown that two cytoskeletal movement proteins, actin (7) and myosin (8), exist in plasmodesmata.
The study of these structures can be aided by appropriate use of computer simulation techniques. This thesis studies in detail one particular set of techniques, that of modelling the dynamic behaviour of the constituent proteins. In this way a deeper understanding of the way in which dynamic structures such as the cytoskeleton are built up and decay, and the way in which material passes through and interacts with (comparatively) large structures such as the plasmodesmata, can be obtained. To a lesser extent, combining such dynamic modelling with other computational techniques such as 3-D modelling and image processing is also explored.
Many of the difficulties faced in examining the cytoskeleton and plasmodesmata can be reduced by using computer analysis in conjunction with experimental techniques. Using computers it is be possible to extract more information out of existing electron micrographs. Computers can also be used to model our theories of cytoskeletal behaviour, and of plasmodesmal structure. Even more exciting is the possibility of using computer modelling to examine the dynamics of cytoskeletal and plasmodesmal function.
To build an accurate model all relevant data must be taken into account. This may be a daunting task, since the amount of possibly applicable data may be huge, so all irrelevant data must simultaneously be excluded. As Albert Einstein said, "Theories should be made as simple as possible, but no simpler".
Deciding what is relevant and what is not is only the first of many difficulties confronting the modeller of complex systems. Often it will be found that it is simply not possible to fully model all factors, and that simplifications and abstractions are necessary to render the problem tractable. This leads to inaccuracies in the model, but may be necessary for the model to be useful. If the inaccuracies become too great, it may be necessary to admit that the system is too complex to model using current techniques and a solution may have to wait on new theoretical and experimental insights, or better computer hardware.
The difficulty with any model is that it is an hypothesis, a theory that the important features of a system can be summarised in a certain way. Thus Newtonian physics, and its derived results, provide an excellent model of, for example, planetary motion. Such model enable the simplification of a vast mass of experimental data into a few simple rules.
Following Karl Popper's observations on theories in general, we can claim that the power of a particular computer model is dependent on the breadth of material it claims to simplify, the strength of its predictions, and the ease with which it makes testable assertions (9). While the computer models developed in this thesis may be interesting, stimulate discussion, and provide useful tools for thinking about nanoscale objects, their ultimate test will be whether they accurately reflect real-world experiment, and can predict the results of new experiments.
 The majority of this thesis is concerned with modelling dynamic
systems, in order to gain insights into complexities of behaviour not
easily modelled with analytic theories. This modelling allows the
investigation of extremely complex systems, with large numbers of
independent proteins interacting.
The majority of this thesis is concerned with modelling dynamic
systems, in order to gain insights into complexities of behaviour not
easily modelled with analytic theories. This modelling allows the
investigation of extremely complex systems, with large numbers of
independent proteins interacting.
In order to establish a baseline, the self assembly of the protein actin (Fig. 1.4), which is well characterised both experimentally and theoretically, is considered in chapter 7, where it is shown how the simulator allows very detailed observation of the assembly process. Continuing on from this work, the self-assembly of the other main cytoskeletal protein, tubulin (which is not so well understood) is covered in Chapter 8.

The simulation engine is then turned to a number of other systems,
such as the self-assembly of protein crystals and viruses (Fig. 1.5).
Some results for these interesting systems are presented in Chapter 9.
There are many other contributions that computers can make to biological analysis. For example, there are a large number of other computational techniques for simplifying data, and for extracting pure data from background noise. One method of particular interest is that of image processing, where the various physical defects or shortcomings of an image can be removed or reduced by computer, revealing details that were not originally fully evident by enhancing otherwise hidden structure. A prime example considered in this thesis is that of electron micrographs, which tend to be indistinct at the level of the cytoskeletal and plasmodesmal structures considered.
The construction of static models can also be an important part of determining structure. Using the information available, it may be possible to clarify the arrangement of components and see what is physically possible, by trying to fit the various pieces together in different ways. Traditionally such models have been either purely mental, sketched (usually on napkins in bars) or made from coloured plasticine and toothpicks.
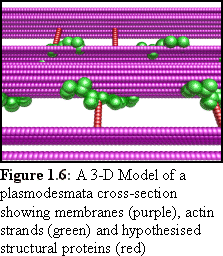 While these are all fine modelling methods, there are some
advantages to using computer modelling as a final stage, and it has
become quite popular in biological research. When it is possible to
accurately specify the sizes of component proteins, it is possible to
work out details such as the size of the largest possible transfer
molecule that can pass through a plasmodesma (the theoretical
"size exclusion limit"). It is possible to readily examine the model
(e.g. Fig. 1.6) from many angles, to see whether it corresponds to
observed real-world objects, and it becomes easier to see whether
hypothesised proteins have sufficient room to exist, and if so, what
configurations are possible.
While these are all fine modelling methods, there are some
advantages to using computer modelling as a final stage, and it has
become quite popular in biological research. When it is possible to
accurately specify the sizes of component proteins, it is possible to
work out details such as the size of the largest possible transfer
molecule that can pass through a plasmodesma (the theoretical
"size exclusion limit"). It is possible to readily examine the model
(e.g. Fig. 1.6) from many angles, to see whether it corresponds to
observed real-world objects, and it becomes easier to see whether
hypothesised proteins have sufficient room to exist, and if so, what
configurations are possible.
As an extension to traditional modelling, this thesis also investigates the possibility of verifying a model by constructing the electron micrograph that such a structure might produce. This "virtual micrograph" allows us to compare the model with actual images of real cellular structures, and thus acts as a validity test.
Results combining the power of the dynamic simulation engine with the above methods of 3-D modelling and 'virtual microscopy' are presented in Chapter 10, along with details of an attempt to improve the 'virtual microscopy' technique by modelling details of the staining and preparation process.
Since this thesis deals with an interdisciplinary topic, an attempt has been made to make it accessible to a wide range of readers. In addition to the main body of the thesis, there are three introductory chapters that attempt to provide a basic understanding of the cell biology, physical chemistry, and computational techniques involved in this work. While the expert may find them superficial or redundant, it is hoped they will provide a minimal background for readers unfamiliar with any or all of these fields, enabling them to follow the work described in the remaining chapters. Readers who are familiar with all three fields may wish to skip these chapters and proceed directly to chapter 5, which is the first to describe work specific to this thesis.
Unfortunately the translation of this thesis from word perfect files to html files had to be done largely by hand. As a result, there may well be occasional problems with references, links and images. The author would be greatful for any such faults to be pointed out. Similarly, if any of the images in the thesis have been incorrectly or inadequately attributed, please contact me so that I can make the necessary alterations!
Index |
Last Chapter |
Next Chapter |
1. Image of cell stained with Coomassie blue from Colin Smith, sourced from Alberts B., et al, (1994) Molecular Biology of the Cell, Garland Publishing, New York, p787
2. Figure from ibid, p962
3. Radford J.E., Vesk, M., and Overall, R.L. (1998) Callose deposition at plasmodesmata, Protoplasma Vol 201 pp 30-37
4. Epel, B.L., van Lent, J.W.M., Cohen, L., Kotlizky, G., Katz, A., and Yahalom, A. (1996) A 41 kDa protein isolated from maize mesocotyl cell walls immunolocalizes to plasmodesmata, Protoplasma Vol 191, pp 70-78
5. Yahalom, A., Lando, R., Katz, A. and Epel, B.L. (1998) A calcium-dependent protein kinase is associated with maize mesocotyl plasmodesmata, J. Plant Phys., Vol 153(3-4) pp 354-362
6. Blackman, L. M., Overall, R. L. (1998) Immunolocalisation of the cytoskeleton to plasmodesmata of Chara corallina, Plant J. Vol 14, pp 733-741.
7. White, R.G., Badelt, K, Overall, R.L. and Vesk, M. (1994) Actin associated with plasmodesmata, Protoplasma Vol 180 pp 169-184
8. Radford, J.E. and White, R.G., (1998) Localization of a myosin-like protein to plasmodesmata, The Plant Journal, Vol 14(6), pp 743-450
9. Popper, K., as expounded over the course of his life, esp in (1934) The Logic of Scientific Discovery.