
Index |
Last Chapter |
Next Chapter |


Felix qui potuit rerum cognoscere causas.
(Happy the man who was able to discover the causes of things.)
___________________________ - Vergil
This chapter presents a very brief guide to the basics of cell function. Readers should be aware that many of the generalised statements presented here are by necessity simplifications and abstractions. Readers interested in a more rigorous treatment are directed to some of the excellent introductory texts available (2)
A summary of biological molecules is given first. These are the building blocks from which the larger cell structures are built, the most important of which are proteins, the ubiquitous 'workhorse' molecule of the cell. Next a small number of larger cell structures are described. These usually consist of different proteins, and the self-assembly of these is described in later chapters. Finally a brief overview of living cells is given, in order to set the context in which the self-assembling structures grow, and to describe the variety of organisms in which they are found.
There are two main types of large molecules, or polymers, within the cell. The first is proteins; which are the workhorses of the cell, the machinery that provides most of the cell's functionality, and makes up most of the structures of the cell. The other type is nucleic acids (such as DNA), which are the data-banks of the cell, containing all the information necessary to build proteins, and to store and pass this information around. This thesis will concern itself mainly with proteins.
There are a variety of types of smaller molecules, or monomers, within the cell. Proteins are almost entirely built from a set of twenty monomers called amino acids. Nucleic acids are made up from a different set of five monomers called nucleotides. The two other main types of small molecules are saccharides (sugars) and fatty acids. Saccharides sometimes polymerise (link together) to make another type of large molecule, the carbohydrates which among other things make up the bulk of the cell walls of plants. (Plant cells, unlike animal cells, have large, thick shells called the "cell wall".)
If proteins are thought of as small machines, amino acids are the nuts and bolts. Amino acids are very small, around ten to thirty atoms. They all have a very similar "head", a carboxyl group, and all save one also have an amino group. The amino groups on each amino acid can easily bind (polymerise) with others, making them easy to string together to form proteins.
A short chain of amino acids is called a peptide, and the chemical covalent bond that joins two amino acids is called a peptide bond. A long string of peptides such as a whole protein or large protein fragment is called a polypeptide.
A protein is more than just a long strip of amino acids however. Most of its interesting properties arise from the overall shape of the entire protein structure. A strip of unrestrained amino acids will coil and fold, since the links between different amino acids are extremely flexible. As it folds, chemical cross links are formed between different parts of the amino acid chain, and in this way it forms its final, stable shape. The process of folding is actually quite complex, and not yet fully understood. In some cases (especially for larger Proteins) other proteins 'help' the Protein to fold to the correct shape.
The unwrapped state, where the protein is an unstructured strip, is known as the denatured form of the protein, which then renatures to the active form, or native state.
Enzymes are special proteins that encourage, or catalyse, specific chemical reactions. The reactions would take place anyway, although in biological terms these reactions might occur so slowly that the organism could have died first. Enzymes make the reactions happen much more quickly. They do this by having binding sites that grab the constituent chemicals to be reacted (also known as substrates). They then use active sites to bring these substrates together to form the final product. Sometimes the enzyme actually changes its shape, or conformation, in order to do this, and sometimes an enzyme needs a helper enzyme called a co-enzyme in order to work.
Most enzymes are named after the name of their product, with the suffix "-ase" added. Hence ribonuclease is an enzyme that acts on ribonucleic acid (RNA), while a protease is something which catalyses the breakdown of proteins in general, and so forth.
Antibodies are small proteins that have a binding site (similar to the binding site on an enzyme) that is designed to link with one particular molecule, such as a specific protein on the outside of a virus. They are very precise, and act as marker flags for the body's immune system; they designate something as being undesirable, for recognition and elimination by white blood cells.
Antibodies are very precise targeting agents, and may entirely ignore a protein with an amino-acid sequence that differs from the target protein by only a single amino acid.
Carbohydrates are sugars, and are used by the cell to store energy. Large, or complex carbohydrates are also used as structural components. Cellulose is one such carbohydrate, which is especially common in the walls of plant cells.
A lipid is a small molecule that is hydrophobic ("water hating") at one end, and occasionally hydrophilic ("water loving") at the other. Pure hydrocarbons (e.g. petrol) are completely hydrophobic, and are insoluble in water, tending to form small isolated drops of oil. Most lipids are like this, but there is a subspecies that lives entirely in membranes, called membrane lipids, and these have both a hydrophilic end and a hydrophobic end, making them amphipathic. As a result, groups of amphipathic lipids tend to orient themselves with their hydrophilic ends placed in the surrounding water, and their hydrophobic ends either embedded in oil, or facing the hydrophobic ends of another group of lipids.
Amphipathic lipids can form sheets. For instance, a planar sheet of lipids might divide a watery volume from an oily volume, having both a hydrophobic surface and a hydrophilic surface. An arrangement that is very common in biology is two such sheets, back to back, called a lipid bilayer. In a lipid bilayer the outside is hydrophilic and the interior is hydrophobic. This is a very stable structure, and lipids of this sort form many of the membranes within cells.
Many processes in the cell require energy to operate. The action of enzymes, the movement of proteins, or the synthesis of DNA and RNA, all require an energy input. The most common method for energy to be delivered is through small molecules such as ATP (adenosine 5'-triphosphate) and GTP (guanosine 5'-triphosphate). The molecules chemically react with the protein they are providing energy to, losing a phosphate ion to become the diphosphate form, ADP or GDP respectively. Many of the experiments and simulations described later in this thesis use large amounts of ATP or GTP to keep reactions energised.
Many of the molecules mentioned above combine to form the large structures found in cells, some of which are visible using a light microscope. Modelling the construction, or self assembly, of some of these structures forms a large part of this thesis.
These membrane-forming lipid bilayers form the cellular sacs that enclose most large cellular components. Cellular components, or organelles, are held together in bags made out of lipid bilayer membranes, and most cells also have a lipid membrane as at least the first layer of the cell boundary. Plant cells, for example, have a lipid bilayer enclosing the cellular liquid, inside the much larger, and leaky, cellulose wall that bounds the cell.
Many proteins are found in or on membranes. Proteins in membranes can usually move around within the membrane fairly freely. Some proteins form small gated pores, which regulate the movement of things from one side of the membrane to the other. Others act to control the mobility of proteins in the membrane, or perform any of the other functions of proteins such as chemical synthesis and transport.

Actin strands are long polymers of the protein actin, often found bundled together in thick strands, webs or gels. They are found in cross-linked networks lying just under the plasma membrane (the outer membrane of the cell), and form the cell cortex.
Actin is notable as a self-assembling protein. A solution of actin monomers in a test tube will, under the right conditions, spontaneously form long actin filaments, which may later break down into single monomers (Fig. 2.1). If energy is provided in the form of free ATP molecules, the cycle may repeat indefinitely.
Actin filaments have a definite directionality, a 'plus' end and a 'minus' end. This directionality can be used by the cell to regulate transport, as the motor proteins that travel along the actin filaments always travel in a specified direction.
Actin is very important in cell movement, and in cell division. A polymerising actin strand can push things along as it extends, such as the pseudo-pod of an amoeba. More usually though, actin serves as a pathway for motor proteins to move along.
Myosin is an important motor protein which, in conjunction with actin, is responsible for much internal cell movement. In animal cells, the actin/myosin complex is responsible for muscular movement, and in plant cells actin and myosin are responsible for cytoplasmic streaming - the stirring of the cell's liquid interior, the cytoplasm, which ensures the mixing of free-floating cell chemicals.

Microtubules grow faster at one end than at the other, and are not usually static. Normally they are either in the process of growing, or the process of shrinking, or growing from one end and shrinking at the other (a feature shared by actin filaments). Associated proteins, known simply as microtubule associated proteins, or MAPs, stabilise the microtubules, and link them to other proteins, to bits of the cell, or to each other.
Microtubules, like actin filaments, are used to transport large proteins and even organelles around. Like actin they have a strong directionality, with a 'plus' end and a 'minus' end. In a cell, many microtubule 'minus' ends meet at a sort of cellular central railway station called a microtubule organising centre, or centrosome. This is usually near the nucleus, with the microtubules streaming away from it toward the cell wall.
Two important microtubule-associated proteins are the motor proteins dynein and kinesin. Dynein links microtubules with other components and moves them towards the minus end, which is often located at a centrosome. The motor protein kinesin moves them away from the minus end (and thus possibly away from a centrosome, towards the periphery of the cell).
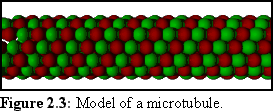
Like actin, tubulin self-assembles from free-floating molecules, and this can be reproduced in a test tube. The structure of a microtubule is also more complex than that of an actin filament, being a number of helices, somewhat like a multi-strand rope. Probably the most common structure is the "3-start, 13-protofilament spiral", which can be thought of as thirteen rows of tubulin monomers joined to make a tube, or equivalently three strands of monomers spiralling to make the same tube (cf Fig. 2.3).
Intermediate filaments are the third main component of the cytoskeleton. They are important structural components and are less dynamic than actin filaments and microtubules. They seem to mainly form structural supports, midway in size between actin and tubulin.
A number of the structures mentioned (especially actin filaments and microtubules) spontaneously self-assemble from their component proteins. An important rate-limiting feature of self-assembly is the need for the structure to reach a minimum size, sometimes called the critical-nucleus, below which it is unstable and quite likely to fall apart, rather than to continue to grow. This process in which a small number of component proteins aggregate to form a critical nucleus is called nucleation, although the term is sometimes applied generically to the rate-limiting step in an assembly process.
Multicellular organisms generally exchange some material between cells, especially in differentiated organisms where distinct parts of the organism, (such as roots and leaves in plants) perform different functions, producing and consuming different material. This material may be food or it may be more complex proteins and hormones. Animal cells have thin cell membranes, and exchange material via small structures called gap junctions which, combined with an elaborate vascular and nervous system, allow material to pass between cells in a controlled manner. Smaller structures called ion channels regulate the flow of ions (very small electrically charged molecules or atoms).
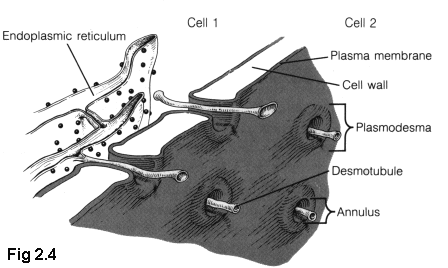
Plant cells have a comparatively thick cellulose cell wall. Although this wall is porous to small molecules, larger molecules require a tunnel of some sort to pass through. Plant cells have such tunnels, called "plasmodesmata" (Fig. 2.4 (5)), but their structure is not well understood, and the examination of this structure is considered in later chapters as an opportunity to apply some of the techniques developed in this thesis.
Although there is some variation in plasmodesmal structure (especially among the "simple" or "lower" plants), generally plasmodesmata have a number of common features. The plasma membrane of a cell extends into the plasmodesmata, forming a protective wall as the outside of the tunnel, separating the interior of the plasmodesmata from the interior of the cell wall. In the very centre of most plasmodesmata is a thin tube of shrunk endoplasmic reticulum (ER), usually called the desmotubule. Between this central pillar and outside wall are a variety of largely unknown proteins (6).
In older plasmodesmata there are often enlargements, where the plasma membrane bulges out into the cell wall space. Sometimes the E.R. of the desmotubule also expands within these cell wall 'bubbles', and sometimes plasmodesmata are observed that meet within the cell wall (although the exact method of formation for these joined or branched plasmodesmata is not fully understood) (7).
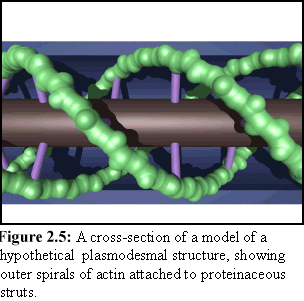
There is still considerable debate about the exact nature of these internal components, and a number of different structural models are considered in Chapter 10. One such model is shown in figure 2.5, which shows the desmotubule as the central gray cylinder, spiral actin strands as green, the external plasma membrane as blue, and hypothesised 'strut' proteins as purple (this particular model is also the basis for the cross-section shown in Fig. 1.6).
While the proteins between the desmotubule and the plasma membrane external wall are not completely known, some recent observations suggest the presence of the cytoskeletal proteins actin (8) and myosin (9) within plasmodesmata, which may imply some continuity between the transport role of the cytoskeleton within the cell, and that of plasmodesmata between cells. Alternatively, this may have more to do with the creation of plasmodesmata, which are formed during cell division. During cell division there are large amounts of cytoskeletal filaments present, which might be caught within forming plasmodesmata. This may even be the way plasmodesmata are formed, by filaments being caught within the growing cell plate that eventually forms a cell wall separating the two sides of the dividing cell (10).
The plasmodesmata transport material between cells. They also have a role in controlling this flow. In extreme cases, such as cell injury, they can close off completely, while there is also evidence to suggest that they partially open and close as part of normal cell operation (11).
In addition to normal cell material being transported, plasmodesmata can also act as a conduit for plant viruses. Some such viruses 'bore' their way through the plasmodesmata, destroying the desmotubule as they go (12), (13) but others seem to be able to pass their genetic material (the RNA or DNA) through without damaging the plasmodesmata (14). Since these molecules are usually much too large to pass through the plasmodesmata, which generally admits only medium-sized molecules, it is thought that the usually highly folded genetic strands must straighten out, and somehow pass through the plasmodesmata like a piece of string through a narrow tube (15).
Living cells are made up of complex combinations of the molecules and structures described here. In increasing order of complexity are viruses (which arguably are not cells at all, since they cannot survive on their own), prokaryotic cells such as bacteria (which have no separate nucleus), and eukaryotic cells (which do have a defined nucleus).
The simplest type of organism is the virus. It is a matter of definitions as to whether it is even alive, or simply a complex protein. At its simplest, it is just a short strand of genetic material (DNA or RNA) enclosed in a shell of protein capsids. If this package finds its way into an appropriate host (a bacterium or a cell) the enclosing shell is broken down, releasing the genetic material, which uses the infected cell to create more capsids and more genetic material, which then in turn self-assembles into more viruses.
More complex are prokaryotic bacteria, simple cells that do not have a separate nucleus. These bacteria developed before nucleated cells, and their genetic material floats inside the same membrane enclosed space as the other cellular organelles. They have been found in rocks over 3 billion years old, which makes them the oldest form of true life. Prokaryotic bacteria are usually simply a loop of DNA in a minimal packet of cell cytoplasm.
Some prokaryotic cells are capable of forming very simple multicellular creatures, such as blue-green algae colonies, but more complex multicelled creatures are all made up from the eukaryotes, the nucleated cells.
These are what we normally think of as a cell. Animal and plant cells are always eukaryotic, meaning that they have a separate membrane-enclosed nucleus containing their DNA. This allows them to regulate the building of proteins more accurately, which in turn allows for far more complex behaviour.
Cells, especially eukaryotic cells, are complex and variant systems. All eukaryotic cells tend to share some common features:
Index |
Last Chapter |
Next Chapter |
1. Images on headpiece courtesy Radford, J.E., Dept. Biological Sciences, Monash University, Melbourne, Australia.
2. Especially recommended are:
Alberts, B.,Bray,D.,Lewis.,J.,Raff,M.,Roberts,K., and Watson J.D., Molecular Biology of the cell (3rd ed.), Garland Publishing, New York,
Lodish, H., Berk, A., Zipursky, S.L., and Matsudaira, P., Molecular Cell Biology (4th ed.) W.H.Freeman & Co., New York, and (slightly more technical)
Voet, D. and Voet, J., Biochemistry, John Wiley & Sons, New York
3. (Detail from) TEM Image from Alberts, B. et al, Molecular Biology of the Cell, (1994), Garland Publishing Inc., New York, Fig. 16-49, p789
4. Li, Qingqin & Joshi, H.C., (1995) -Tubulin Is a Minus End-specific Microtubule Binding Protein, J. Cell. Biol., Vol 131, No 1., pp 207-214.
5. Image from Taiz, L. and Zeigler, E. (1991) Plant Physiology, Benjamin/Cummings Publishing Company, inc., California.
6. Epel, B.L, (1994) Plasmodesmata: Composition, Structure and Trafficking, Plant Mol.Biol., Vol 26, pp 1343-1356
7. Lucas, W.J., Ding, B., Van der Schoot, C. (1993) Plasmodesmata and the supracellular nature of plants New Phytol. Vol 125, pp 435-476
8. White, R.G. et al. (1994) op. cit.
9. Radford, J.E. et al. (1998) op. cit.
10. Hepler, P.K., and Bonsignone, C.L. (1990) Caffeine inhibition of cytokinesis: ultrastructure of cell plate formation/degraadation, Protoplasma Vol 157 pp182-192
11. Epel, B.L. op. cit.
12. Van Lent, J., Storms, M., Van der Meer, F., Wellink, J., and Goldbach, R. (1991) Tubular structures involved in movement of cowpea mosaic virus are also formed in infected cowpea protolasts., J. Gen. Virol. Vol 72, pp 2615-2623
13. Kim, K.S., and Lee K. W. (1992) Geminivirus induced macrotubules and their suggested role in cell-to-cell movement. Phyto.Path. Vol 82 pp 664-669
14. Wolf,S., Deom, C.M., Beachy, R.N. and Lucas, W.J. (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit Science Vol 246, pp 377-379.
15. Ghoshory, S., Lartey,R., Sheng, J., and Citovsky, V. (1997) Transport of proteins and nucleic acids through plasmodesmata Ann. Rev. Plant Phys. Plant Mol. Biol. Vol 48: pp 27-50